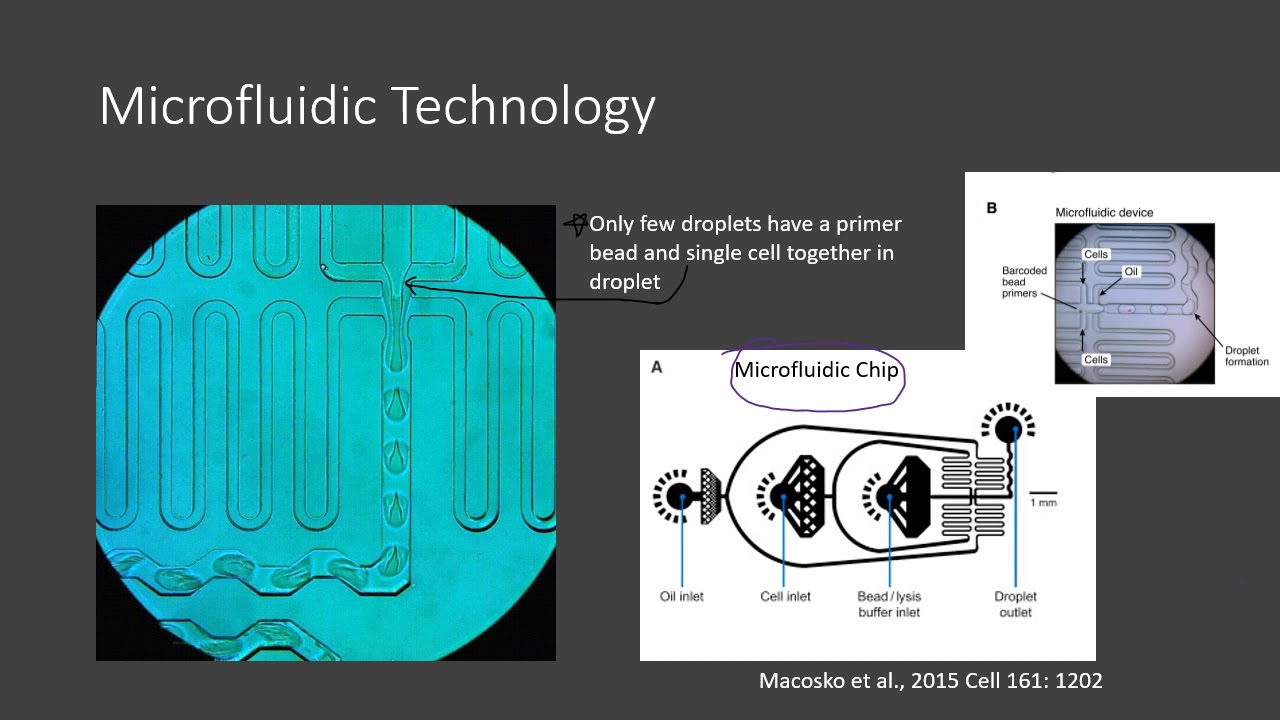
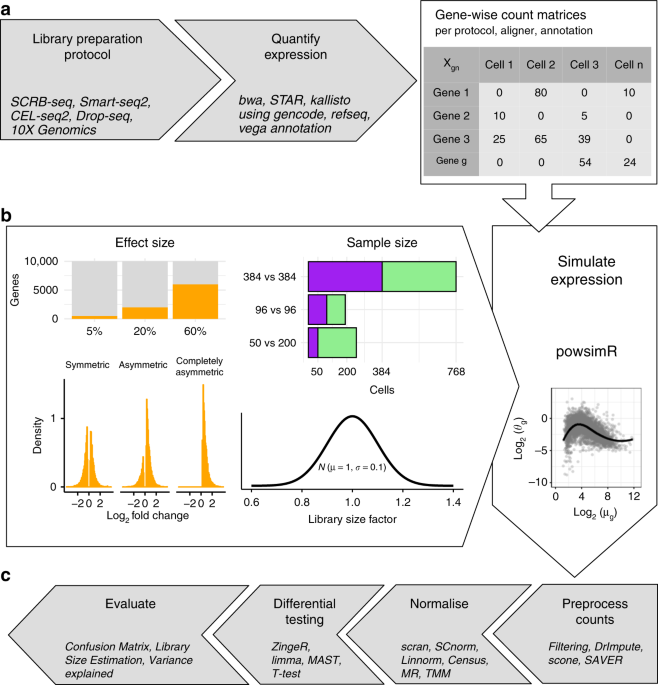
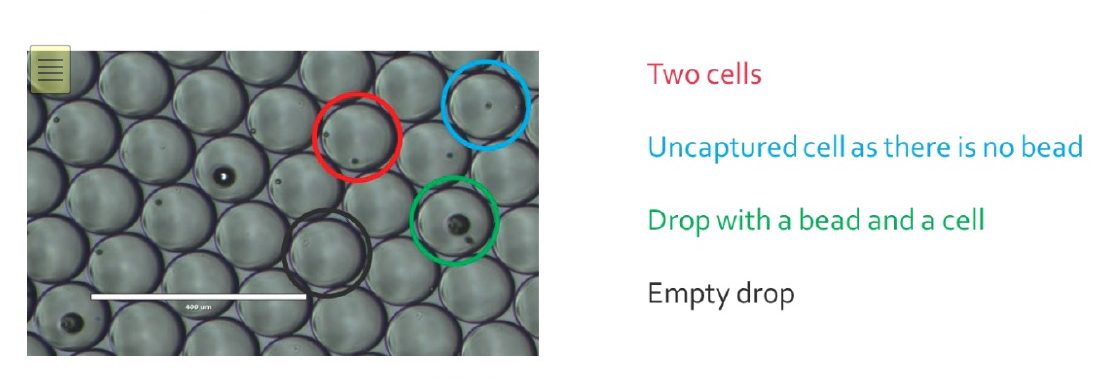
Comparative analysis of NovaSeq 6000 and MGISEQ 2000 single‐cell RNA sequencing data - Chen - 2022 - Quantitative Biology - Wiley Online Library
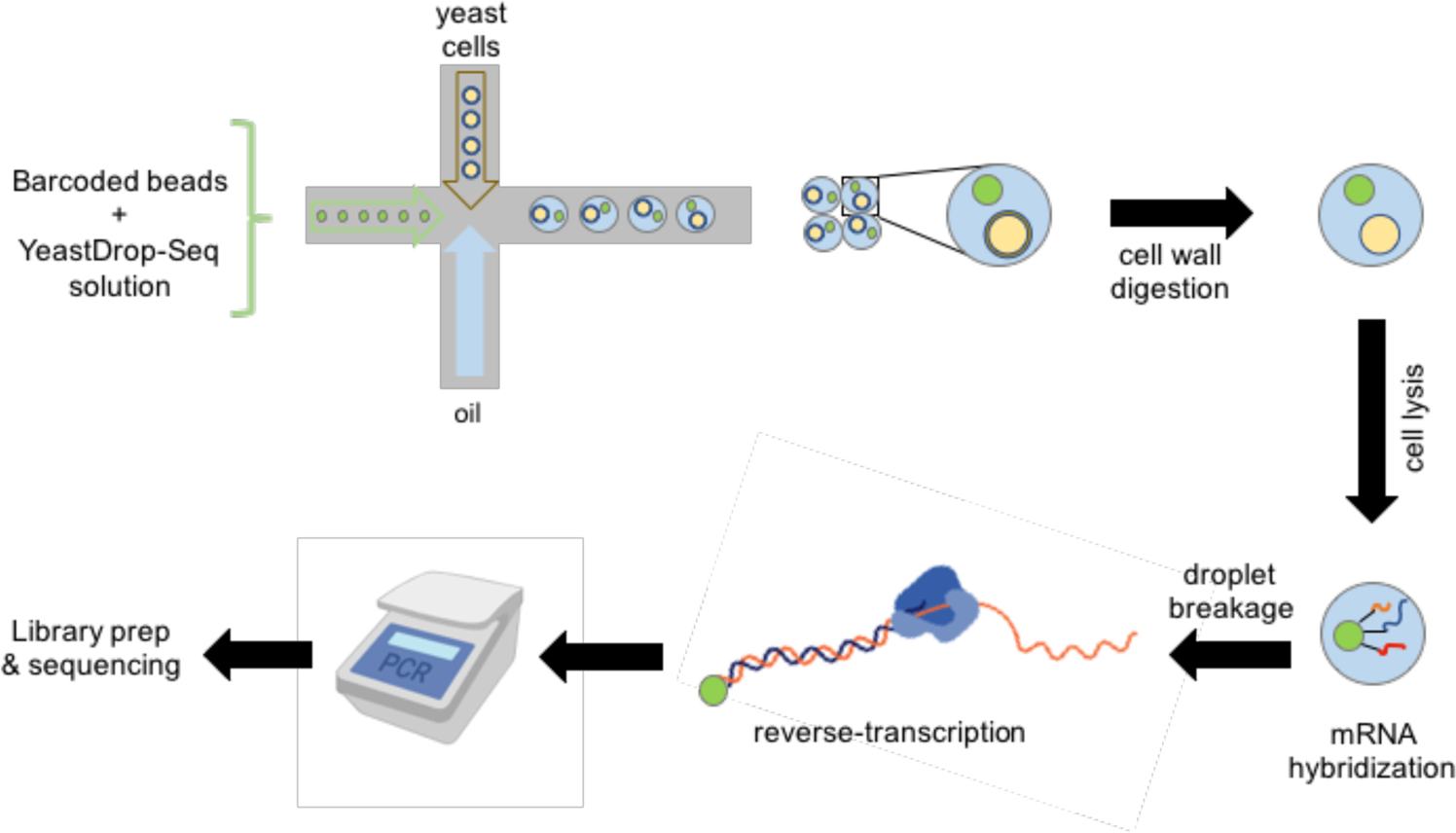
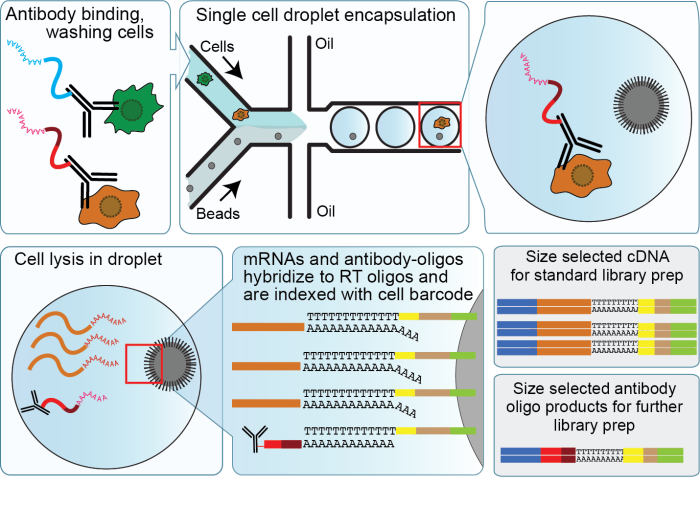
A yeast-optimized single-cell transcriptomics platform elucidates how mycophenolic acid and guanine alter global mRNA levels | Communications Biology

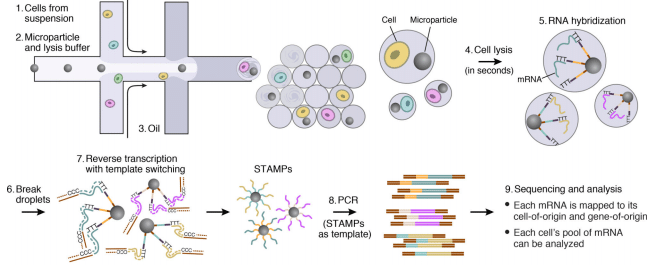
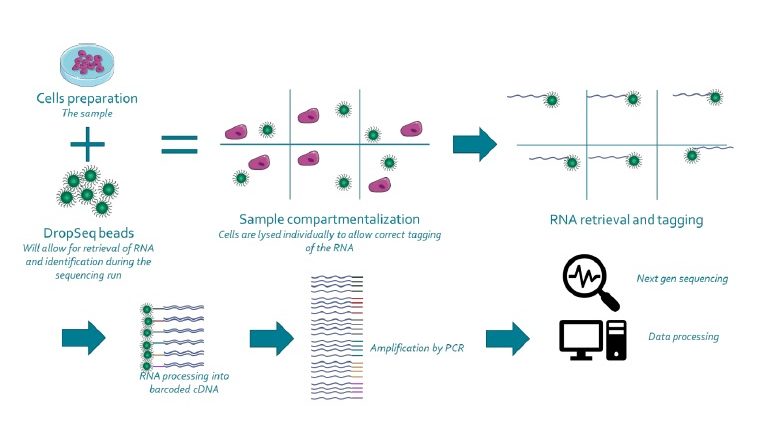
Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets - ScienceDirect
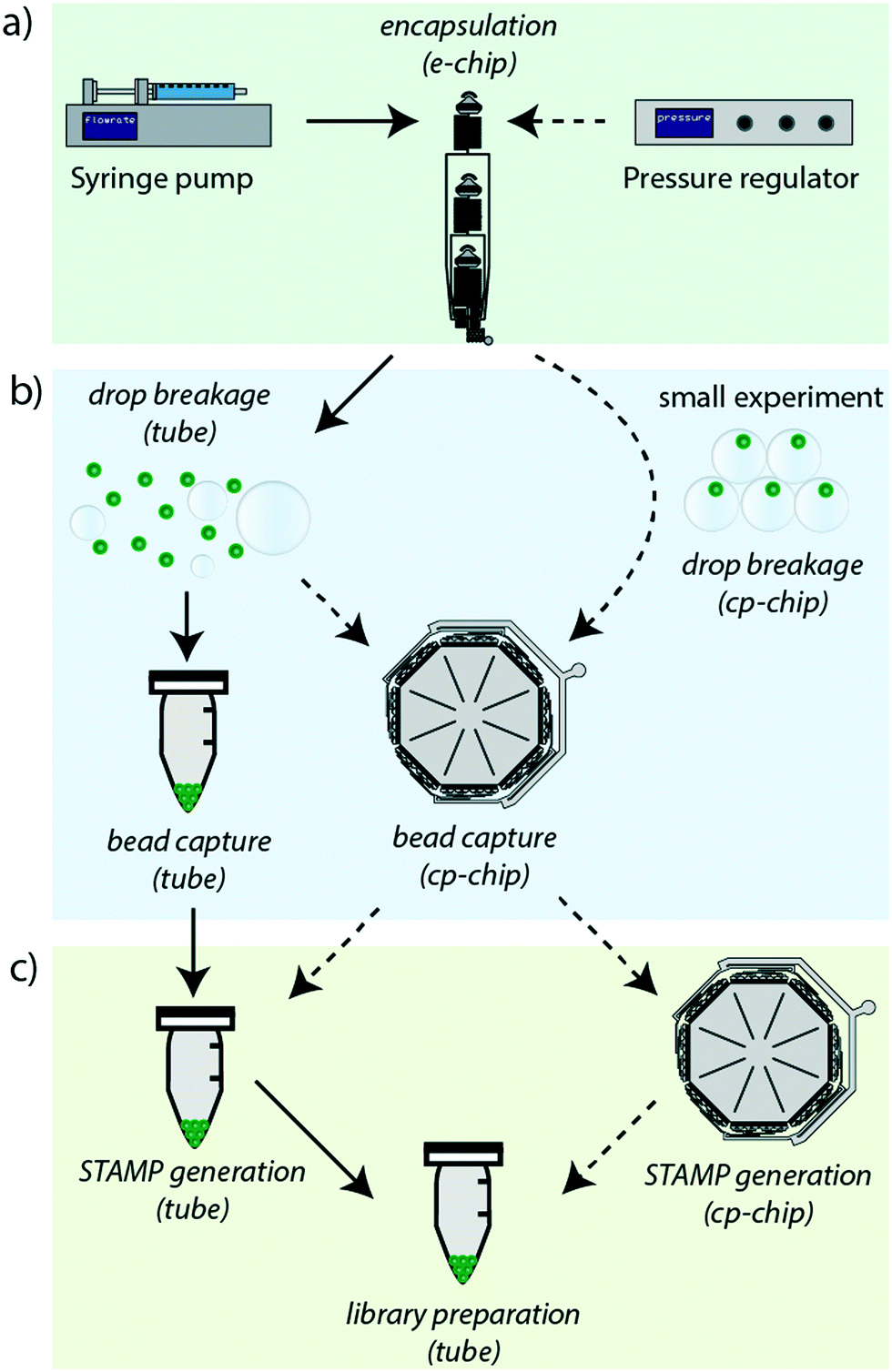
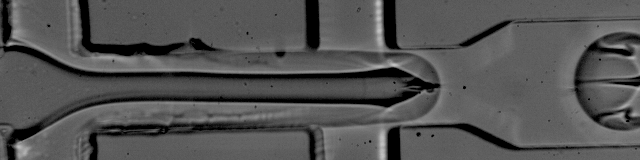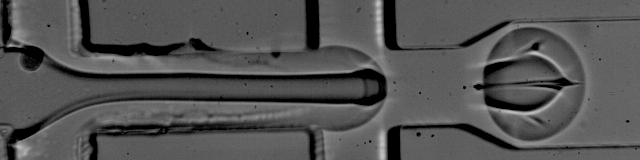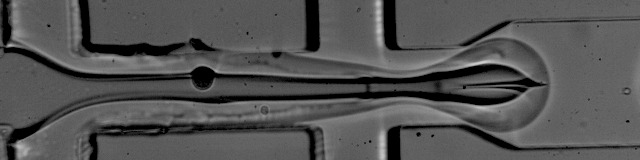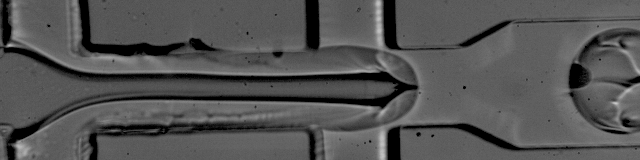
Simplified Drop-seq workflow with minimized bead loss using a bead capture and processing microfluidic chip - Lab on a Chip (RSC Publishing) DOI:10.1039/C9LC00014C